Getting Started with SURF-SHYFEM¶
This guide provides instructions for downloading, installing, and running SURF-SHYFEM in a virtual machine (VM) environment. It complements the video tutorials available on the SURF website and the instructions provided in the SURF-NEMO manual.
Get the Surf Virtual Machine¶
- Download VirtalBox following the SURF-NEMO manual
-
Download the SURF VM from the SURF webpage.
-
Refer to the SURF-NEMO manual to correctly setup the VM. Some additional notes:
- The login username and passwords are surf and surf2020 respectively.
- If you use a AZERTY keyboard, you should type the password as if you were using a QWERTY keyboard (i.e. don't press shift when entering the numbers). You'll be able to switch to AZERTY once inside the VM, from the system settings.
- To run the Madagascar test case, you must resize the virtual machine. To do that: shut down the virtual machine, go to the folder containing the
.vdifile and type in a Terminal:Then start the virtual machine, go toVBoxManage modifyhd surf_scratch.vdi --resize 60000administration → GParted → devsdband resize. - You may need to modify the VM storage unit. By default, it points to
/Users/Franz/VirtualBox VMs/Surf/VBoxGuestAdditions.iso. You should redirect it to theVBoxGuestAdditions.isodisk image available from your VirtualBox installation (e.g.usr/share/VirtualBox/VBoxGuestAdditions.isofor linux systems).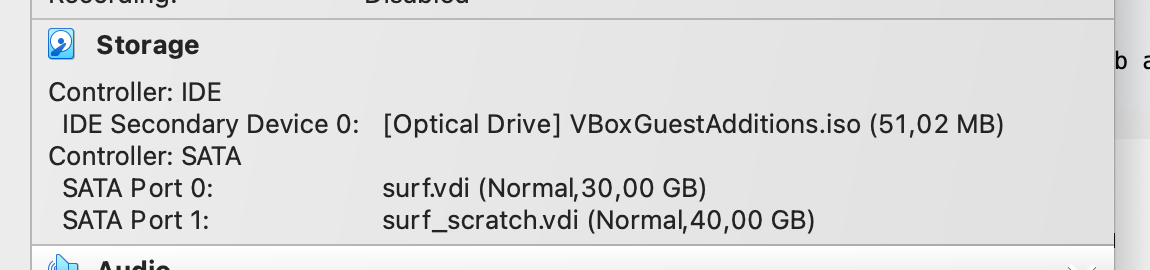
Installing SHYFEM¶
-
Start the virtual machine.
-
To download
surf_datasetandsurf_shyfemzip files, open a Terminal and type:cd /scratch/surf/surf_install/releases/ echo "Downloading SURF-Datasets..." wget https://www.surf-platform.org/repository/surf_datasets/surf_datasets_1.01/surf_datasets_1.01.tar.gz wait echo "Downloading SURF-SHYFEM..." wget https://www.surf-platform.org/repository/surf_shyfem/surf_shyfem_1.00/surf_shyfem_1.00.tar.gz wait -
From the same location, install the datasets and SHYFEM:
These commands install the surf static datasets and SURF-Shyfem underinstall.sh surf_dataset_1.01 install.sh surf_shyfem/scratch/surf/surf_datasets/and/scratch/surf/surf_shyfem/, respectively. -
Once done, you must grant execution permissions to script files:
chmod -R a+x /scratch/surf/surf_shyfem/surf_shyfem_1.00/
Run the Surf-SHYFEM Testcase (Madagascar coastal area)¶
-
Download the testcase input datasets from the SURF webpage:
cd /scratch/surf/indata_offline/ wget https://www.surf-platform.org/repository/surf_shyfem/surf_shyfem_1.00/case_studies/madagascar_20210402_indata.tar.gz tar -zxvf madagascar_20210402_indata.tar.gz -
Create the experiment directory and move the data in it:
mkdir -p /scratch/surf/experiments/mdg_etoofs/data/indata/ cp -r /scratch/surf/indata_offline/madagascar_20210402_indata/madagascar_20210402/* /scratch/surf/experiments/mdg_etoofs/data/indata/ -
Activate virtual environment:
conda activate surf_shyfem -
You're all set.
Unstructured grid generation¶
The first step to the execution of a downscaling experiment is the creation of the unstructured grid in the downscaled domain (please refer to the video tutorials for a comprehensive introduction on the subject).
-
Generate the grid with the embedded tool (based on Gmsh):
Note:cd /scratch/surf/surf_shyfem/current/grid-tool/ python create_grid.py @ci osm @cb 49.4682 -12.0557 49.3572 -12.1062 49.2196 -12.1272 49.1467 -12.3619 49.3724 -12.4503 49.5137 -12.3577 @ob 49.5652,-11.9743 49.7047,-12.1988- The commas are only in the @ob section. The syntax is:
python create_grid.py @ci osm @cb lon1 lat1 lon2 lat2 ... @ob lon1,lat1 lon2,lat2 ... - Coordinates must be written in decimal notation
- Both
@cband@obcoordinates must be in an anticlockwise sense
- The commas are only in the @ob section. The syntax is:
-
Follow on-screen prompts to set the grid parameters (for their description please refer to the video tutorials):
You can play a bit with the grid parameters and bounding coordinates, but at first we advise to stick with these values to get the first experiment done.Insert L_open: 0.05 Insert L_coast: 0.01 Insert Delta_open: 0.015 Insert Delta_coast: 0.002 -
Move the grid to the correct directory and rename it:
Manually editmkdir /scratch/madagascar cp /scratch/surf/surf_shyfem/current/grid-tool/output/* /scratch/madagascar mv /scratch/madagascar/new.grd /scratch/madagascar/madagascar_final.grd/scratch/madagascar/madagascar_final.grdto remove the first blank line.
Run the downscaling experiment¶
-
Create a new folder in the directory /scratch/from_GUI/ and let's call it
mdg_etoofs(this is the Experiment ID), and copy to it the input configuration files:mkdir /scratch/from_GUI/mdg_etoofs cp /scratch/surf/surf_shyfem/surf_shyfem_1.00/input_param* /scratch/from_GUI/mdg_etoofs/ -
Copy the
surf_shyfemsource code to the experiment directory:ln -s /scratch/surf/surf_shyfem/current/* /scratch/surf/experiments/mdg_etoofs -
Run the experiment:
cd /scratch/surf/surf_shyfem/current/scripts; python create_exp.py mdg_etoofs
Known issues¶
-
You have to remove the first blank line from the file
File "/scratch/surf/experiments/mdg_etoofs_ext/code/ocean/scripts/bathyI nterp/shyfem.py", line 28, in inBox idNodes = self.nodes[:, 1].astype(int) IndexError: too many indices for array: array is 1-dimensional, but 2 were indexednew.grd. -
```bash warning : nodes in element *** are in clockwise sense ``` Apply ```/scratch/surf/surf_shyfem/current/shyfem/fembin/exgrd -a``` to your input `new.grd` file. If it does not work, change the order of nodes in the `new.grd` file by hand. -
```bash FileNotFoundError: [Errno 2] No such file or directory: b'/scratch/surf/experiments/mdg_prova1/data/indata/ocean/20210402_d -MERCATOR--RFVL--MDG-b20210413_an-fv01.nc' ``` Manually insert in `/scratch/surf/experiments/{experiment_name}/indata/` the input data for ocean atmosphere and observations needed for your experiment. -
```bash STOP error stop adjust_levels: hlv too low ``` Select correctly the levels in `input_param_basic.json`. Your modeled ocean is not deep enough. -
Did you correctly select the path in
STOP error stop (<something about boundaries>)boundariesPathofinput_param_basic.jsonfiles? -
If the simulation breaks when reading the initial conditions files, check if there is a blank line somewhere. E.g., for
STOP error stop iff_inituvin.dat, there may be one before “v-velocity”. You cannot remove it, since SURF will create the file again when launching the simulation. A workaround (ninja skills required) is to manually stop (ctrl+Z) thecreate_exp.pyexecution right after the creation of the problematic file (sayuvin.dat), manually remove the blank line that is causing the error and then resume the process execution (fg %[job ID]) -
Good question. There should be a previous error, so you should look above in the error traceback to fix it.
mv: cannot stat 'out.nc': No such file or directory -
There is a failure in the final plots generation. This is expected, since the postprocessing tools are not yet stable. If the files
fatal:MapRGDHDrawMapList: MDRGSF/MDRGOF - ERROR OPENING RANGS/GSHHS CAT FILE fatal:PlotManagerPreDraw: error in plot pre-draw.nos.ncand.ous.nchave been produced, the experiment has correctly concluded.